Protein Dynamical Structure Revealed by Temperature-jump Time-resolved Infrared Spectroscopy
Date:08-05-2014 Print
Protein could carry out its biological function only when it is folded into the right conformation, and the conformation might be changing during the activation process, therefore protein dynamical structure provides a key clue to understand its function. Although many protein static structures have been resolved by X-ray crystallography, 2D NM and cryoelectron microscope at atomic level, the static structure would be different from the dynamical structure, especially under physiological condition.
In their continuous effort to study the protein dynamical structure with temperature-jump time-resolved infrared spectroscopy (Biophys. J. 2007, 93, 2756-2766; Biophys. J. 2009, 97, 2756-2766), a team lead by Prof. WENG Yuxiang from the Institute of Physics (IOP), Chinese Academy of Sciences recently collaborated with Prof. CHANG Zengyi’s team in Peking University to investigate the dynamical structure of the E. coil DegP involving in the first step of its activation. They found that the disassembly of the DegP hexamer follows a “proteinquake” manner, as shown in the figure below, such that the sequential unfolding/disassembly process initiated by the interfacial β-sheets serving as the temperature sensor and epicenter finishes within about 134 ns at room temperature.
The E. coil DegP is a representative of the heat shock protein family. It has been reported to possess both chaperone and protease activities. When in a resting state, DegP exists as a hexamer consisting of two trimeric units linked by interfacial βstrands with its protease active sites being blocked. It has been shown that upon substrate binding, DegP hexamers can be transformed into the active cage-like 12- or 24-mers with the substrate being included inside the cage. It is believed that the first step for the activation of DegP hexamer would involve its disassembly into two trimmers. In addition, the protease activity of DegP increases when temperature rises in a certain range. Therefore, an important question arises that whether the activation of DegP hexamer is triggered by the substrate or the temperature.
The critical point in their work is to correctly assign the IR finger prints of the secondary structural components, and the assignments were confirmed by a number of DegP mutants. They found that the interfacial secondary structural components have degreed thermal stability, and finally revealed the detailed dynamics for disassembly DegP hexamer into trimmers occurring in a proteinquake mode, which eventually leads to the dissociation of the two trimeric units. Remarkably, the disassembly process occurs even at the room temperature (25℃), demonstrating that temperature is not the key factor that triggers the activation process of DegP.
This work demonstrates again that the temperature-jump time-resolved infrared spectroscopy can be an effective method for investigating the correlation between the biological function and structure for large protein molecules.
This work has been published recently on Scientific Reports [Sci. Rep. 4, 4834 (2014)].
This work was supported by the Natural Science Foundation of China (Grant No. 20925313 and 21090342), National Basic Research Program of China (Grant No. 2006CB910302) and Chinese Academy of Sciences Innovation Program (Grant No. KJCX2-YW-W25).
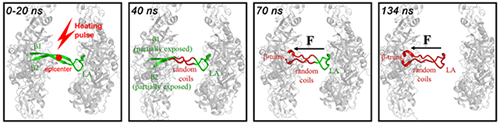 |
| Fig 1. Schematic illustration of the proteinquake mechanism for the thermal-induced dissociation of the DegP hexamer with four typical snapshots at the indicated time points. Only one (green colored) of the six β1-LA-β2 structures at the interface of the trimeric units is displayed. The unfolded structures are red colored. The epicenter (denoted as a red circle) locates at the fully exposed part of the interfacial β-sheet. F denotes the force of the tension. The black arrows denote the direction of the motion of the LA loop. (Image by WENG Yuxiang et al.) |


