Optimal targeting of cells
Date:27-06-2017 Print
A key challenge in biomedical research is the ability to specifically target cells and tissues. Cells can often be recognised by the concentrations of receptors expressed on their surface. A good targeting strategy results in strong binding to cells with the desired receptor profile, and no binding (or very weak binding) to other cells.
Targeted drug delivery is usually based on identifying a specific marker (peptide, sugar) that is unique to the targeted group of cells. Binding to a single marker type can be effective if this molecule is presented in sufficient quantities on the outer surface of the targeted cell. However, in many cases of practical importance (e.g., many types of cancer), the markers that are known are not unique to cancer cells, but just overexpressed. Over the past 20 y many nanoparticle-based targeting methods have been developed. However, thus far, effective tumor drug delivery is hampered by the lack of reliable, unique markers.
It is clearly important to exploit all of the information that we have about the concentration of various receptors on the cell surface and then design guest particles that target this specific receptor profile. To recognize the simultaneous presence of a mixture of different receptors on the host surface, we need to use a “guest” particle (e.g., drug-delivery vehicle) that is coated with a mixture of cognate ligands schematically shown in Fig. 1.
 |
|
Fig.1 Schematic diagram of the problem (a). Nanoparticle (NP) is targeting a membrane with several types of receptors(represented as concave shapes). The main challenge is how to design the ligand composition on the particle to achieve maximum selectivity, e.g. (b) to target cell B but not cell A or cell C. |
Researchers from Institute of Physics, Dr. Tine Curk lead by the group leader Prof. Jure Dobnikar (SM-9), together with a long-term collaborator Prof. Daan Frenkel from University of Cambridge, used computer simulations and statistical physics to formulate a theory for optimal multivalent targeting. The key findings are summarized as simple design rules for multivalent particles distinguishing target cells based on their receptor profile:
- the concentration profiles on the guest and target surface should be density matched;
- the effective strength of the ligand-receptor interaction should be about 1.3 kBT.
The design rules discovered in this work are conceptually different from the traditional approaches in biomedicine, which are based on identifying a marker (highly-expressed receptor), and choosing a ligand (antibody) that strongly and specifically binds to this marker. The antibodies cannot be selective: they strongly attach to the target cells but also to other cells with similar membrane composition. As a result, in cancer treatment the drugs are delivered to both, the cancer cells and healthy cells (e.g. liver, kidney…), which strongly limits the efficiency of chemotherapy. Here it is shown that high selectivity can only be achieved by multivalent carriers with weak bonds. Using the above results, it is possible to design targeted drug delivery with substantially reduced side effects.
The study entitled “Optimal multivalent targeting of membranes with many distinct receptors” was published in PNAS: http://www.pnas.org/content/early/2017/06/20/1704226114. The study was supported by the Chinese Academy of Sciences, Institute of Physics and grants from Europe and UK.
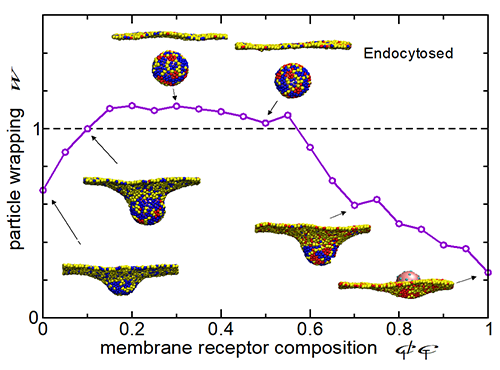 |
| Fig.2 The rate of endocytosis of NPs through the cell membrane can be controlled by the design of particle’s surface profile.. |
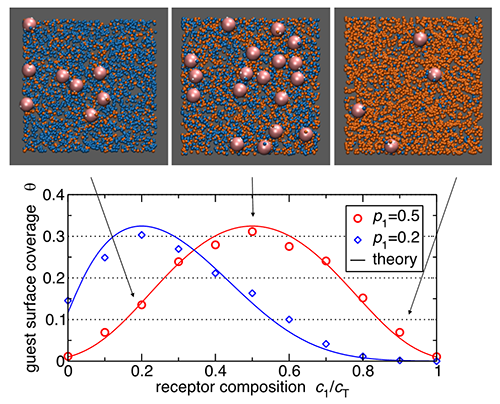 |
| Fig.3 Nanoparticle binding to cell membranes is controlled by receptor composition. |
Contact:
Institute of Physics
DOBNIKAR Jure
Email:jd489@cam.ac.uk
and
CURK Tine
Email:tc@iphy.ac.cn
Key words:
multivalency | Monte Carlo simulations | drug delivery | endocytosis | statistical mechanics
Abstract:
A key challenge in biomedical research is the ability to specifically target cells and tissues. We show that properly designed multivalent targeting of multiple receptor types results in a specificity toward a chosen receptor density profile. Thus, demonstrating a general route toward targeting cells without particularly dominant markers.


