New method speeds up identification of viruses and bacteria
Date:07-04-2020 Print
The current coronavirus crisis highlights this need for fast and accurate detection infections. Both viral infections like coronavirus and bacterial infections can be detected by screening for DNA in patient samples. But this is challenging because the amount of disease DNA is small and it has to be detected in the presence of other, non-disease, DNA. There is a great need to improve the sensitivity of the current methods and to design simple and reliable ways to detect DNA of interest in the presence of other DNA.
The standard approach is to design molecular probes that bind strongly to the disease DNA but not to the non-disease DNA. In the new study we used computer simulations to demonstrate how this could be done better. The idea is that instead of designing molecular probes that bind strongly to one place on the target DNA, we should, counterintuitively, design probes that bind weakly all over the target DNA and exploit the concept of super-selectivity to realize selective multivalent binding of the probes to the target DNA. We have developed a numerical scheme to identify optimal probe sequences and tested our approach in large-scale, coarse-grained simulations. The designed probes can indeed distinguish between viral and bacterial genome, and even between two different strands of E. coli bacterium.
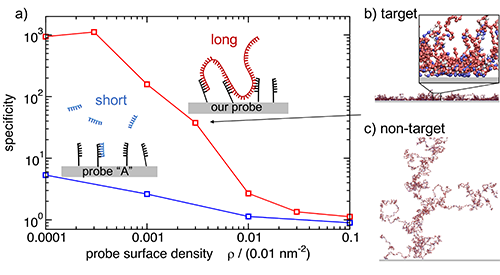 |
| Simulating multivalent detection of bacterial DNA.(a)Specificity of binding of target vs non-target DNA to surfaces coated with probes targeting E. coli. The blue curve shows results for a published probe, and the red curve for our top-scoring multivalent probe binding.(b-c)Snapshots from our simulations for genomic DNA of E. coli(b)and B. subtilis(c)binding to the surface coated in multivalent probes. The blow-up section in (b) shows that blue blobs with a stronger surface-binding interaction are predominantly found close to the surface. Source: PNAShttps://doi.org/10.1073/pnas.1918274117. |
This work opens up new possibilities to construct robust and cost-efficient methods to detect infectious diseases. Given the urgent need for fast, reliable disease detection methods, it is expected to have a strong impact and to encourage new experimental work leading to development of new health care products.
The study entitled Computational design of probes to detect bacterial genomes by multivalent binding was published in the Proceedings of the National Academy of Sciences of the USA. The work coordinated by Rosalind J. Allen was performed within a multinational team of researchers in UK, China and Slovenia, with a crucial involvement of IoP: besides the SM9 group members Tine Curk, James D. Farrell and Jure Dobnikar, regular visiting fellows Daan Frenkel, Erika Eiser and Stefano Angioletti-Uberti were involved.
Publication:
T. Curk, C. A. Brackley, J. D. Farrell, Z. Xing, D. Joshi, S. Direito, U. Bren, S. Angioletti-Uberti, J. Dobnikar, E. Eiser, D. Frenkel, R. J. Allen, Computational design of probes to detect bacterial genomes by multivalent binding, accepted for publication, in press, Proc. Nat. Ac. Sci. USA (2020), https://doi.org/10.1073/pnas.1918274117
Contact:
Institute of Physics, CAS
Jure Dobnikar
Jd489@cam.ac.uk
Keywords:
DNA-based detection, multivalent binding, super-selectivity, computer simulations, polymer physics
Abstract:
Rapid methods for diagnosis of bacterial infections are urgently needed to reduce inappropriate use of antibiotics, which contributes to antimicrobial resistance. In many rapid diagnostic methods, DNA oligonucleotide probes, attached to a surface, bind to specific nucleotide sequences in the DNA of a target pathogen. Typically, each probe binds to a single target sequence, i.e., target-probe binding is monovalent. Here we show using computer simulations that the detection sensitivity and specificity can be improved by designing probes that bind multivalently to the entire length of the pathogen genomic DNA, such that a given probe binds to multiple sites along the target DNA. Our results suggest that multivalent targeting of long pieces of genomic DNA can allow highly sensitive and selective binding of the target DNA, even if competing DNA in the sample also contains binding sites for the same probe sequences. Our results are robust to mild fragmentation of the bacterial genome. Our conclusions may also be relevant for DNA detection in other fields, such as disease diagnostics more broadly, environmental management and food safety.


