Scientists reveal how SID-1 recognizes dsRNA and initiates systemic RNAi
Date:26-04-2024 Print
RNA interference (RNAi) is a fascinating biological process in worms, plants, fungi, and metazoans that has been used as a valuable tool in studying gene functions and as therapeutics. In Caenorhabditis elegans, the multipass transmembrane protein, systemic RNA interference defective protein 1 (SID-1), has an indispensable role in the uptake and spreading of double-stranded RNA (dsRNA) between cells and tissues, resulting in systemic RNAi. In addition, two human SID-1 homologs, SID1 transmembrane family member 1 (SIDT1) and SIDT2, were suggested to be involved in RNA transport.
However, the underlying molecular mechanisms of how SID-1 specifically distinguishes dsRNA from single-stranded RNA (ssRNA) and DNA and facilitates subsequent dsRNA transport between cells remain unknown. The answers to those questions would be important for understanding systemic RNAi and aiding in RNA-related applications. Recently, Dr. Zhang Jiangtao in Prof. JIANG Daohua’s group from the Institute of Physics, Chinese Academy of Sciences, by combining cryo-EM, in vitro, and in vivo experiments, demonstrated how SID-1 specifically recognizes dsRNA and provided important insights into the internalization of dsRNA by SID-1.
For over two decades, SID-1 was thought to act as a dsRNA channel. Here, the researchers solved high-resolution cryo-EM structures of SID-1 and the human SID-1 homologs SIDT1 and SIDT2 (Fig. 1a), which revealed the conserved architecture of C. elegans and human SID-1 homologs. The SID-1 homologs are organized in a homo-dimeric fashion. Surprisingly, the SID-1 dimer does not show any obvious pore within the transmembrane domain (TMD), suggesting that SID-1 may not function as a dsRNA channel. MST binding assays confirmed that SID-1 can potently and specifically bind to dsRNA but not dsDNA.
Subsequently, the researchers obtained the cryo-EM structure of SID-1–dsRNA complex (Fig. 1b), demonstrating the detailed dsRNA binding mode and the molecular determinants for how SID-1 distinguishes dsRNA from ssRNA and DNA. Interestingly, such determinants are not present in human SIDT1 or SIDT2. The structural findings were supported by mutagenesis studies using MST binding assays, dsRNA uptake in S2 cells, and in vivo systemic RNAi assays.
Finally, the researchers show that the removal of the long intracellular loop transmembrane helices 1 and 2 did not affect SID-1 dimerization, cell localization, or dsRNA binding, but it significantly impaired dsRNA uptake in S2 cells (Fig. 2a) and systemic RNAi in C. elegans (Fig. 2b). Moreover, co-localization revealed that SID-1 and dsRNA co-locate in vesicle-like subcellular organelles (Fig. 2c). Based on these results, the researchers propose that SID-1 functions as a dsRNA receptor and facilitates subsequent dsRNA internalization via recruiting endocytic related proteins using the long loop (Fig. 2d).
This study entitled “Structural insights into double-stranded RNA recognition and transport by SID-1” was published on Nature Structural & Molecular Biology.
DOI : 10.1038/s41594-024-01276-9
The study was supported by the National Science Foundation of China, Institute of Physics, and the Chinese Academy of Sciences.
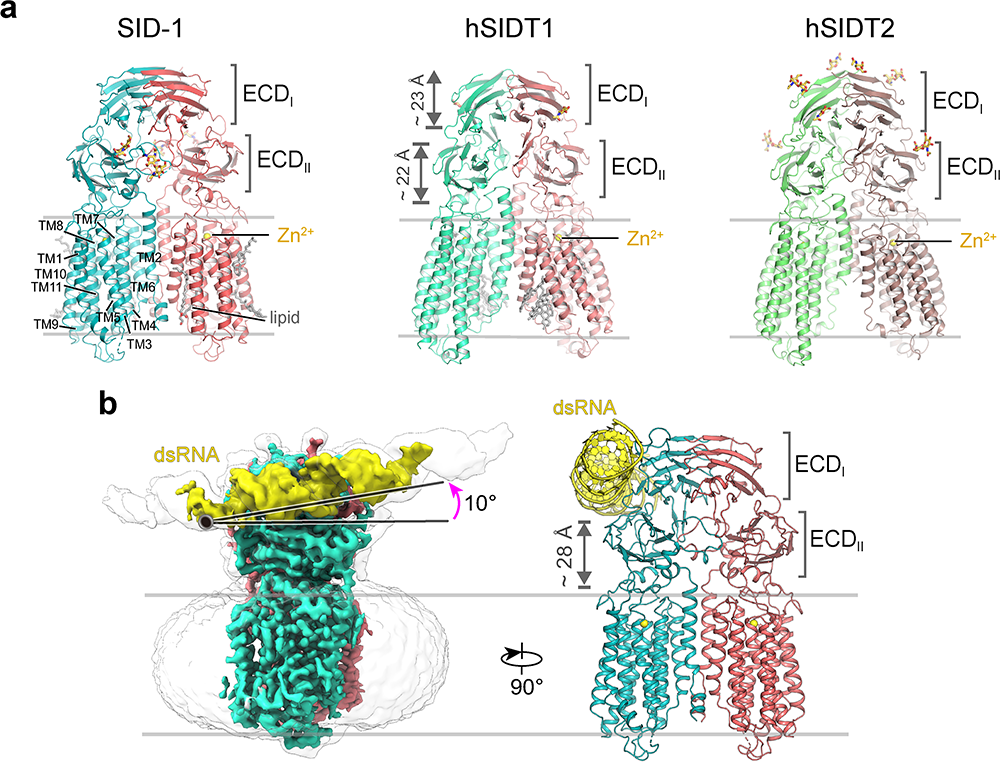
Fig.1 Cryo-EM structures of SID-1 homologs (a) and the SID-1/dsRNA complex (b). (Image by the Institute of Physics)
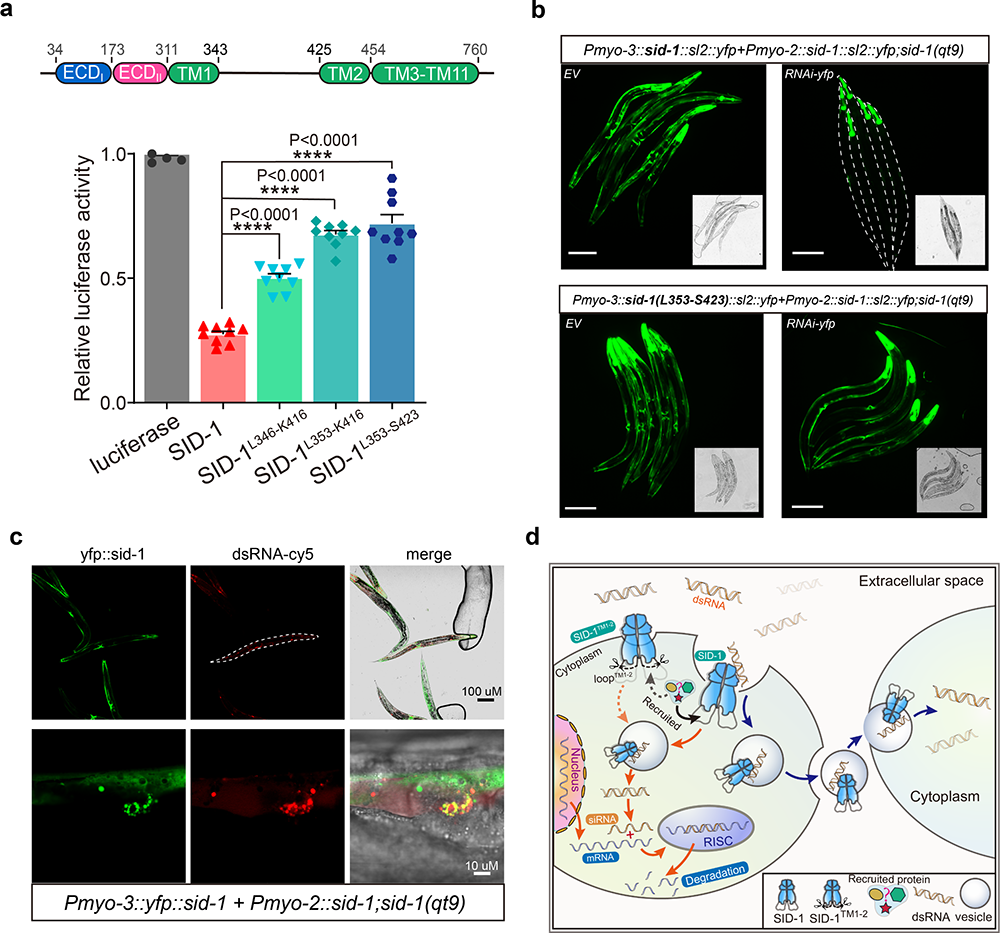
Fig.2 a. dsRNA uptake in S2 cells of SID-1TM1-2 mutants; b. Defects in systemic RNAi of SID-1TM1-2; c. The colocalization of SID-1 and dsRNA; d. The proposed model of the internalization of dsRNA by SID-1.
Contact:
Institute of Physics
JIANG Daohua
Email:jiangdh@iphy.ac.cn
Key words:
systemic RNA interference; SID-1 homologs; Cryo-EM; dsRNA receptor
Abstract:
RNA uptake by cells is critical for RNA-mediated gene interference (RNAi) and RNA-based therapeutics. In Caenorhabditis elegans, RNAi is systemic as a result of SID-1-mediated dsRNA across cells. Despite the functional importance, the underlying mechanisms of dsRNA internalization by SID-1 remain elusive. Here, we describe cryo-EM structures of SID-1, SID-1/dsRNA complex, human SID-1 homologs SIDT1 and SIDT2, elucidating the structural basis of dsRNA recognition and import by SID-1. The homo-dimeric SID-1 homologs share conserved architecture, but only SID-1 possesses the molecular determinants within its extracellular domains for distinguishing dsRNA from single-stranded RNA and DNA. We show that the removal of the intracellular loop between transmembrane helix 1 and 2 attenuates the dsRNA uptake and the systemic RNAi in vivo, suggesting a possible endocytic mechanism of SID-1 mediated dsRNA internalization. Our study provides mechanistic insights into dsRNA internalization by SID-1, which may facilitate the development of dsRNA applications based on SID-1.


