XPR1 transport and regulation mechanisms revealed by cryo-EM
Date:22-08-2024 Print
Phosphorus is an essential element for all domains of life and is involved in bone and tooth growth, biogenesis of lipids and nucleic acids, high-energy organic compound metabolism, and protein signaling. Cellular inorganic phosphate (Pi) levels are tightly regulated, and imbalance of Pi homeostasis leads to severe human diseases. Xenotropic and polytropic retrovirus receptor 1 (XPR1) is the only known Pi exporter in mammals to date, loss-of-function mutations of XPR1 are associated with human neurological disease primary familial brain calcification (PFBC) and renal Fanconi syndrome. XPR1 consists of a transmembrane domain (TMD) and a soluble SPX domain, which can sense intracellular InsPP levels and regulates XPR1 activity.
However, the architecture of XPR1 and how the SPX domain communicates with the TMD remain largely unknown. Recently, Prof. Jiang Daohua's group revealed the cryo-electron microscopy (cryo-EM) structures of human XPR1 in three distinct functional states with Pi and inositol hexakisphosphate (InsP6)-bound, demonstrating the structural features and Pi export mechanism of XPR1-like proteins.
Here, the researchers determined high-resolution cryo-EM structures of closed, open and InsP6-bound states. The XPR1 structure is organized in a homo-dimeric manner (Fig.1). On the basis of the structural and functional results, the authors proposed a model for Pi transport and regulation by the SPX domain in XPR1 homologs (Fig.2). The bound Pi in Site 2 interacts with W573 and triggers local conformational changes in TM9b which opens the gate domain. In addition, the SPX domain binds to InsP6 and facilitates Pi efflux by liberating the C-terminal loop that limits Pi entry.
This study elucidates a transport mechanism that XPR1 resembles a channel-like gating mechanism rather than the canonical alternating-access mechanism for most transporter proteins, and explains the mode of action of how the SPX domain senses InsPPs and regulates Pi efflux. In conclusion, the novel transport mechanism of the coupling between InsPPs sensing and Pi export in XPR1 significantly expand our understanding of cellular regulation of Pi homeostasis.
This study entitled “Human XPR1 structures reveal phosphate export mechanism” was published on Nature. Link for the article:https://www.nature.com/articles/s41586-024-07852-9。
This study was supported by the National Natural Science Foundation of China, The Chinese Academy of Sciences and The Institute of Physics.
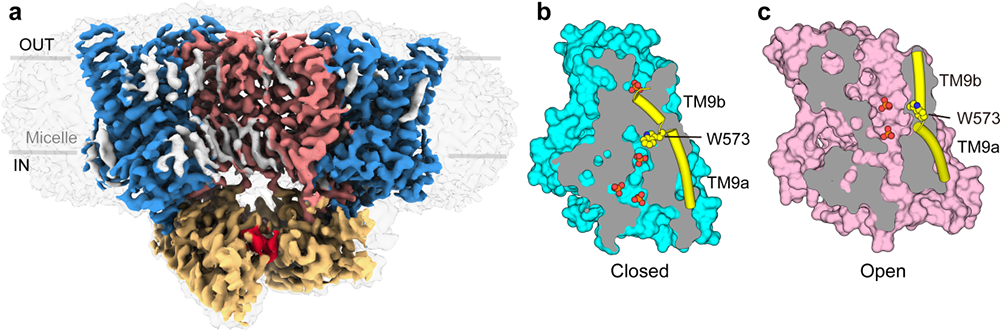
Fig.1 Cryo-EM structures of XPR1 in InsP6-bound, closed and open states (Image from Institute of Physics, SM10)
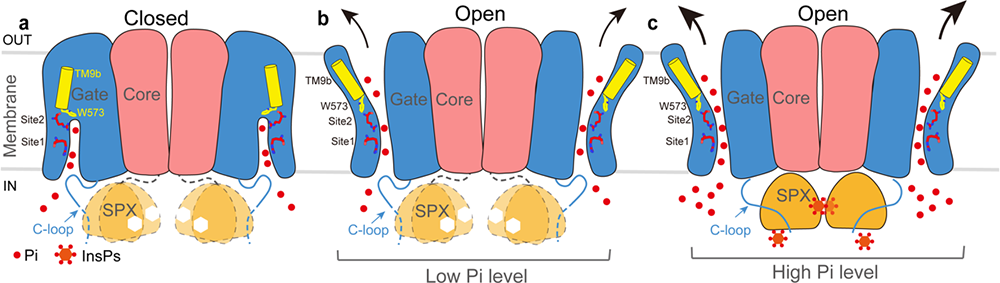
Fig.2 Working model for Pi efflux by XPR1 (Image from Institute of Physics, SM10)
Contact:
Institute of Physics, Laboratory of Soft Matter Physics.
Jiang Daohua
Email: jiangdh@iphy.ac.cn.
Key words:
XPR1; The SPX domain; Cryo-EM; Transport mechanism; Phosphate homeostasis.
Abstract:
Inorganic phosphate (Pi) is a fundamental macronutrient for all living organisms, the homeostasis of which is critical for numerous biological activities1-3. As the only known human Pi exporter to date, XPR1 plays an indispensable role in cellular Pi homeostasis4,5. Dysfunction of XPR1 is associated with neurodegenerative disease6-8. However, the mechanisms underpinning XPR1-mediated Pi efflux and regulation by the intracellular inositol polyphosphate (InsPP) sensor SPX domain remain poorly understood. Here, we present cryo-EM structures of human XPR1 in Pi-bound closed, open, and InsP6-bound forms, revealing the structural basis for XPR1 gating and regulation by InsPPs. XPR1 consists of an N-terminal SPX domain, a dimer-formation core domain and a Pi transport domain. Within the transport domain, three basic clusters are responsible for Pi binding and transport, and a conserved Trp573 acts as a molecular switch for gating. In addition, the SPX domain binds to InsP6 and facilitates Pi efflux by liberating the C-terminal loop that limits Pi entry. This study provides a conceptual framework for the mechanistic understanding of Pi homeostasis by XPR1 homologs in fungi, plants, and animals.


